Posterior Predictive Checks
In this tutorial you will learn how to run posterior predictive checks in HDDM.
A posterior predictive check is a very useful tool when you want to evaluate if your model can reproduce key patterns in your data. Specifically, you can define a summary statistic that describes the pattern you are interested in (e.g. accuracy in your task) and then simulate new data from the posterior of your fitted model. You can the apply the the summary statistic to each of the data sets you simulated from the posterior and see if the model does a good job of reproducing this pattern by comparing the summary statistics from the simulations to the summary statistic caluclated over the model.
What is critical is that you do not only get a single summary statistic from the simulations but a whole distribution which captures the uncertainty in our model estimate.
Lets do a simple analysis using simulated data. First, import HDDM.
import hddm
Simulate data from known parameters and two conditions (easy and hard).
data, params = hddm.generate.gen_rand_data(params={'easy': {'v': 1, 'a': 2, 't': .3},
'hard': {'v': 1, 'a': 2, 't': .3}})
First, lets estimate the same model that was used to generate the data.
m = hddm.HDDM(data, depends_on={'v': 'condition'})
m.sample(1000, burn=20)
Next, we’ll want to simulate data from the model. By default,
post_pred_gen() will use 500 parameter values from the posterior
(i.e. posterior samples) and simulate a different data set for each
parameter value.
ppc_data = hddm.utils.post_pred_gen(m)
The returned data structure is a pandas DataFrame object with a
hierarchical index.
ppc_data.head(10)
| rt | response | |||
|---|---|---|---|---|
| node | sample | |||
| wfpt(easy) | 0 | 0 | 0.41009 | 1 |
| 1 | 0.79089 | 1 | ||
| 2 | -0.67769 | 0 | ||
| 3 | 0.49359 | 1 | ||
| 4 | 1.59039 | 1 | ||
| 5 | 0.99669 | 1 | ||
| 6 | 5.51089 | 1 | ||
| 7 | 0.73069 | 1 | ||
| 8 | 0.82829 | 1 | ||
| 9 | 0.92839 | 1 |
The first level of the DataFrame contains each observed node. In
this case the easy condition. If we had multiple subjects we would get
one for each subject.
The second level contains the simulated data sets. Since we simulated 500, these will go from 0 to 499 – each with generated from a different parameter value sampled from the posterior.
The third level is the same index as used in the data and numbers each trial in your data.
For more information on how to work with hierarchical indices, see the Pandas documentation.
There are also some helpful options like append_data you can pass to
post_pred_gen().
help(hddm.utils.post_pred_gen)
Help on function post_pred_gen in module kabuki.analyze:
post_pred_gen(model, groupby=None, samples=500, append_data=False, progress_bar=True)
Run posterior predictive check on a model.
:Arguments:
model : kabuki.Hierarchical
Kabuki model over which to compute the ppc on.
:Optional:
samples : int
How many samples to generate for each node.
groupby : list
Alternative grouping of the data. If not supplied, uses splitting
of the model (as provided by depends_on).
append_data : bool (default=False)
Whether to append the observed data of each node to the replicatons.
progress_bar : bool (default=True)
Display progress bar
:Returns:
Hierarchical pandas.DataFrame with multiple sampled RT data sets.
1st level: wfpt node
2nd level: posterior predictive sample
3rd level: original data index
:See also:
post_pred_stats
Now we want to compute the summary statistics over each simulated data
set and compare that to the summary statistic of our actual data by
calling post_pred_stats().
ppc_compare = hddm.utils.post_pred_stats(data, ppc_data)
print ppc_compare
observed mean std SEM MSE credible \
stat
accuracy 0.890000 0.874580 0.063930 0.000238 0.004325 1
mean_ub 1.084831 1.048314 0.111169 0.001334 0.013692 1
std_ub 0.654891 0.542704 0.129186 0.012586 0.029275 1
10q_ub 0.510200 0.549030 0.045206 0.001508 0.003551 1
30q_ub 0.649200 0.704437 0.067714 0.003051 0.007636 1
50q_ub 0.818000 0.891622 0.099117 0.005420 0.015244 1
70q_ub 1.253800 1.165027 0.149496 0.007881 0.030230 1
90q_ub 1.884400 1.741424 0.282329 0.020442 0.100152 1
mean_lb -0.970818 -1.046499 0.269939 0.005728 0.078595 1
std_lb 0.543502 0.423627 0.251797 0.014370 0.077772 1
10q_lb 0.547000 0.660271 0.203289 0.012830 0.054157 1
30q_lb 0.648000 0.785022 0.223779 0.018775 0.068852 1
50q_lb 0.693000 0.939898 0.269239 0.060958 0.133448 1
70q_lb 1.022000 1.158692 0.346341 0.018685 0.138637 1
90q_lb 1.666000 1.532752 0.515372 0.017755 0.283363 1
quantile mahalanobis
stat
accuracy 55.500000 0.241202
mean_ub 64.699997 0.328490
std_ub 81.500000 0.868420
10q_ub 18.900000 0.858949
30q_ub 20.700001 0.815736
50q_ub 23.400000 0.742775
70q_ub 73.500000 0.593812
90q_ub 71.500000 0.506417
mean_lb 57.517658 0.280362
std_lb 72.754791 0.476077
10q_lb 26.538849 0.557192
30q_lb 25.933401 0.612307
50q_lb 14.228052 0.917022
70q_lb 40.060543 0.394675
90q_lb 65.893036 0.258547
As you can see, we did not have to define the summary statistics as by
default, HDDM already calculates a bunch of useful statistics for RT
analysis such as the accuracy, mean RT of the upper and lower boundary
(ub and lb respectively), standard deviation and quantiles. These are
listed in the rows of the DataFrame.
For each distribution of summary statistics there are multiple ways to
compare them to the summary statistic obtained on the observerd data.
These are listed in the columns. observed is just the value of the
summary statistic of your data. mean is the mean of the summary
statistics of the simulated data sets (they should be a good match if
the model reproduces them). std is a measure of how much variation
is produced in the summary statistic.
The rest of the columns are measures of how far the summary statistic of
the data is away from the summary statistics of the simulated data.
SEM = standard error from the mean, MSE = mean-squared error,
credible = in the 95% credible interval.
Finally, we can also tell post_pred_stats() to return the summary
statistics themselves by setting call_compare=False:
ppc_stats = hddm.utils.post_pred_stats(data, ppc_data, call_compare=False)
print ppc_stats.head()
accuracy mean_ub std_ub 10q_ub 30q_ub 50q_ub \
(wfpt(easy), 0) 0.96 1.164858 0.825420 0.500940 0.736800 0.909940
(wfpt(easy), 1) 0.92 1.066229 0.500696 0.552553 0.725853 0.842753
(wfpt(easy), 2) 0.84 1.106792 0.660981 0.538767 0.708527 0.852747
(wfpt(easy), 3) 0.90 0.949962 0.524693 0.507878 0.634398 0.784638
(wfpt(easy), 4) 0.88 0.967202 0.523246 0.509131 0.638661 0.781231
70q_ub 90q_ub mean_lb std_lb 10q_lb 30q_lb \
(wfpt(easy), 0) 1.388660 1.902310 -1.270140 0.592450 0.796180 1.033160
(wfpt(easy), 1) 1.298903 1.815803 -0.921803 0.204067 0.720703 0.842803
(wfpt(easy), 2) 1.137547 1.791117 -1.610109 1.577114 0.813307 0.843867
(wfpt(easy), 3) 1.013418 1.533458 -1.125698 0.371009 0.667518 1.004518
(wfpt(easy), 4) 0.958081 1.826761 -0.765531 0.363230 0.545531 0.599181
50q_lb 70q_lb 90q_lb
(wfpt(easy), 0) 1.270140 1.507120 1.744100
(wfpt(easy), 1) 0.899403 0.964963 1.140823
(wfpt(easy), 2) 1.084597 1.183127 2.654677
(wfpt(easy), 3) 1.334438 1.347158 1.454438
(wfpt(easy), 4) 0.614681 0.665531 1.136381
This DataFrame has a row for each simulated data set. The columns
are the different summary statistics.
Defining your own summary statistics
You can also define your own summary statistics and pass them to
post_pred_stats():
ppc_stats = hddm.utils.post_pred_stats(data, ppc_data, stats=lambda x: np.mean(x), call_compare=False)
ppc_stats.head()
| stat | |
|---|---|
| (wfpt(easy), 0) | 1.067459 |
| (wfpt(easy), 1) | 0.907187 |
| (wfpt(easy), 2) | 0.672088 |
| (wfpt(easy), 3) | 0.742396 |
| (wfpt(easy), 4) | 0.759274 |
Note that stats can also be a dictionary mapping the name of the
summary statistic to its function.
Using PPC for model comparison with the groupby argument
One useful application of PPC is to perform model
comparison. Specifically, you might estimate two models, one for which
a certain parameter is split for a condition (say drift-rate v for
hard and easy conditions to stay with our example above) and one in
which those conditions are pooled and you only estimate one
drift-rate.
You then want to test which model explains the data better to assess
whether the two conditions are really different. To do this, we can
generate data from both models and see if the pooled model
systematically misses aspects of the RT data of the two
conditions. This is what the groupby keyword argument is
for. Without it, if you ran post_pred_gen() on the pooled model
you would get simulated RT data which was not split by
conditions. Note that while the RT data will be split by condition,
the exact same parameters are used to simulate data of the two
conditions as the pooled model does not separate them. It simply
allows us to match the two conditions present in the data to the
jointly simulated data more easily.
m_pooled = hddm.HDDM(data) # v does not depend on conditions
m_pooled.sample(1000, burn=20)
ppc_data_pooled = hddm.utils.post_pred_gen(m_pooled, groupby=['condition'])
You could then compare ppc_data_pooled to ppc_data above (by
passing them to post_pred_stats) and find that the model with
separate drift-rates accounts for accuracy (mean_ub) in both
conditions, while the pooled model can’t account for accuracy in
either condition (e.g. lower MSE).
Summary statistics relating to outside variables
Another useful way to apply posterior predictive checks is if you have
trial-by-trial measure (e.g. EEG brain measure). In that case the
append_data keyword argument is useful.
Lets add a dummy column to our data. This is going to be uncorrelated to anything but you’ll get the idea.
from numpy.random import randn
data['trlbytrl'] = randn(len(data))
m_reg = hddm.HDDMRegressor(data, 'v ~ trlbytrl')
m_reg.sample(1000, burn=20)
ppc_data = hddm.utils.post_pred_gen(m_reg, append_data=True)
from scipy.stats import linregress
ppc_regression = []
for (node, sample), sim_data in ppc_data.groupby(level=(0, 1)):
ppc_regression.append(linregress(sim_data.trlbytrl, sim_data.rt_sampled)[0]) # slope
orig_regression = linregress(data.trlbytrl, data.rt)[0]
plt.hist(ppc_regression)
plt.axvline(orig_regression, c='r', lw=3)
plt.xlabel('slope')
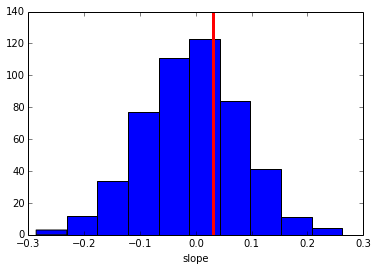
As you can see, the simulated data sets have on average no correlation to our trial-by-trial measure (just as in the data) but we also get a nice sense of the uncertainty in our estimation.