Fitting go/no-go using the chi-qsuare approach
by Jan Willem de Gee (jwdegee@gmail.com)
This is a demo of fitting go/no-go data with HDDM using the chi-qsuare method, as described in
de Gee JW, Tsetsos T, McCormick DA, McGinley MJ & Donner TH. 2018. Phasic arousal optimizes decision computations in mice and humans. bioRxiv. (https://www.biorxiv.org/content/early/2018/10/19/447656).
See also
Ratcliff, R., Huang-Pollock, C., & McKoon, G. (2016). Modeling individual differences in the go/no-go task with a diffusion model. Decision, 5(1), 42-62 (http://psycnet.apa.org/record/2016-39470-001).
# Load packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import hddm
from joblib import Parallel, delayed
Let’s start with defining some functionality
def get_choice(row):
if row.condition == 'present':
if row.response == 1:
return 1
else:
return 0
elif row.condition == 'absent':
if row.response == 0:
return 1
else:
return 0
def simulate_data(a, v, t, z, dc, sv=0, sz=0, st=0, condition=0, nr_trials1=1000, nr_trials2=1000):
"""
Simulates stim-coded data.
"""
parameters1 = {'a':a, 'v':v+dc, 't':t, 'z':z, 'sv':sv, 'sz': sz, 'st': st}
parameters2 = {'a':a, 'v':v-dc, 't':t, 'z':1-z, 'sv':sv, 'sz': sz, 'st': st}
df_sim1, params_sim1 = hddm.generate.gen_rand_data(params=parameters1, size=nr_trials1, subjs=1, subj_noise=0)
df_sim1['condition'] = 'present'
df_sim2, params_sim2 = hddm.generate.gen_rand_data(params=parameters2, size=nr_trials2, subjs=1, subj_noise=0)
df_sim2['condition'] = 'absent'
df_sim = pd.concat((df_sim1, df_sim2))
df_sim['bias_response'] = df_sim.apply(get_choice, 1)
df_sim['correct'] = df_sim['response'].astype(int)
df_sim['response'] = df_sim['bias_response'].astype(int)
df_sim['stimulus'] = np.array((np.array(df_sim['response']==1) & np.array(df_sim['correct']==1)) + (np.array(df_sim['response']==0) & np.array(df_sim['correct']==0)), dtype=int)
df_sim['condition'] = condition
df_sim = df_sim.drop(columns=['bias_response'])
return df_sim
def fit_subject(data, quantiles):
"""
Simulates stim-coded data.
"""
subj_idx = np.unique(data['subj_idx'])
m = hddm.HDDMStimCoding(data, stim_col='stimulus', split_param='v', drift_criterion=True, bias=True, p_outlier=0,
depends_on={'v':'condition', 'a':'condition', 't':'condition', 'z':'condition', 'dc':'condition', })
m.optimize('gsquare', quantiles=quantiles, n_runs=8)
res = pd.concat((pd.DataFrame([m.values], index=[subj_idx]), pd.DataFrame([m.bic_info], index=[subj_idx])), axis=1)
return res
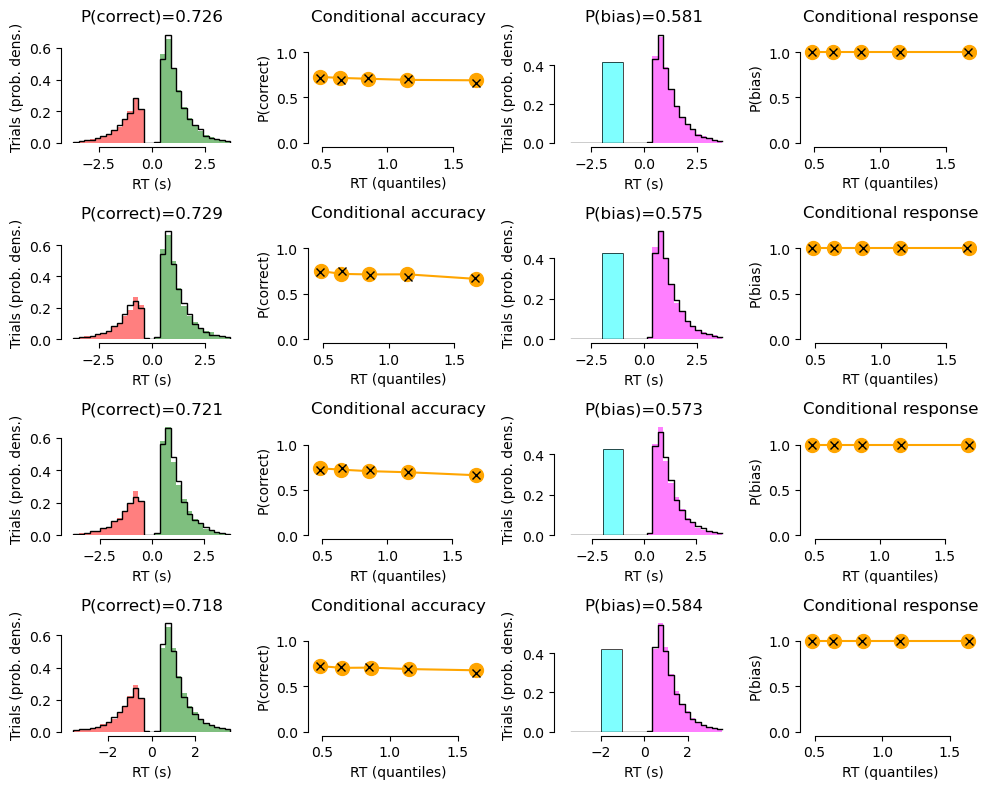
def summary_plot(df_group, df_sim_group=None, quantiles=[0, 0.1, 0.3, 0.5, 0.7, 0.9,], xlim=None):
"""
Generates a
"""
nr_subjects = len(np.unique(df_group['subj_idx']))
fig = plt.figure(figsize=(10,nr_subjects*2))
plt_nr = 1
for s in np.unique(df_group['subj_idx']):
df = df_group.copy().loc[(df_group['subj_idx']==s),:]
df_sim = df_sim_group.copy().loc[(df_sim_group['subj_idx']==s),:]
df['rt_acc'] = df['rt'].copy()
df.loc[df['correct']==0, 'rt_acc'] = df.loc[df['correct']==0, 'rt_acc'] * -1
df['rt_resp'] = df['rt'].copy()
df.loc[df['response']==0, 'rt_resp'] = df.loc[df['response']==0, 'rt_resp'] * -1
df_sim['rt_acc'] = df_sim['rt'].copy()
df_sim.loc[df_sim['correct']==0, 'rt_acc'] = df_sim.loc[df_sim['correct']==0, 'rt_acc'] * -1
df_sim['rt_resp'] = df_sim['rt'].copy()
df_sim.loc[df_sim['response']==0, 'rt_resp'] = df_sim.loc[df_sim['response']==0, 'rt_resp'] * -1
max_rt = np.percentile(df_sim.loc[~np.isnan(df_sim['rt']), 'rt'], 99)
bins = np.linspace(-max_rt,max_rt,30)
# rt distributions correct vs error:
ax = fig.add_subplot(nr_subjects,4,plt_nr)
N, bins, patches = ax.hist(df.loc[:, 'rt_acc'], bins=bins,
density=True, color='green', alpha=0.5)
for bin_size, bin, patch in zip(N, bins, patches):
if bin < 0:
plt.setp(patch, 'facecolor', 'r')
if df_sim is not None:
ax.hist(df_sim.loc[:, 'rt_acc'], bins=bins, density=True,
histtype='step', color='k', alpha=1, label=None)
ax.set_title('P(correct)={}'.format(round(df.loc[:, 'correct'].mean(), 3),))
ax.set_xlabel('RT (s)')
ax.set_ylabel('Trials (prob. dens.)')
plt_nr += 1
# condition accuracy plots:
ax = fig.add_subplot(nr_subjects,4,plt_nr)
df.loc[:,'rt_bin'] = pd.qcut(df['rt'], quantiles, labels=False)
d = df.groupby(['rt_bin']).mean().reset_index()
ax.errorbar(d.loc[:, "rt"], d.loc[:, "correct"], fmt='-o', color='orange', markersize=10)
if df_sim is not None:
df_sim.loc[:,'rt_bin'] = pd.qcut(df_sim['rt'], quantiles, labels=False)
d = df_sim.groupby(['rt_bin']).mean().reset_index()
ax.errorbar(d.loc[:, "rt"], d.loc[:, "correct"], fmt='x', color='k', markersize=6)
if xlim:
ax.set_xlim(xlim)
ax.set_ylim(0, 1.25)
ax.set_title('Conditional accuracy')
ax.set_xlabel('RT (quantiles)')
ax.set_ylabel('P(correct)')
plt_nr += 1
# rt distributions response 1 vs 0:
ax = fig.add_subplot(nr_subjects,4,plt_nr)
if np.isnan(df['rt']).sum() > 0:
# some initial computations
bar_width = 1
fraction_yes = df['response'].mean()
fraction_yes_sim = df_sim['response'].mean()
no_height = (1 - fraction_yes) / bar_width
no_height_sim = (1 - fraction_yes_sim) / bar_width
hist, edges = np.histogram(df.loc[:, 'rt_resp'], bins=bins, density=True,)
hist = hist * fraction_yes
hist_sim, edges_sim = np.histogram(df_sim.loc[:, 'rt_resp'], bins=bins, density=True,)
hist_sim = hist_sim * fraction_yes_sim
# Add histogram from go choices
# ground truth
ax.bar(edges[:-1], hist, width=np.diff(edges)[0], align='edge',
color='magenta', alpha=0.5, linewidth=0,)
# simulations
ax.step(edges_sim[:-1] + np.diff(edges)[0], hist_sim, color='black', lw=1)
# Add bar for the no-go choices (on the negative rt scale)
# This just illustrates the probability of no-go choices
# ground truth
ax.bar(x=-1.5, height=no_height, width=bar_width, alpha=0.5, color='cyan', align='center')
# simulations
ax.hlines(y=no_height_sim, xmin=-2, xmax=-1, lw=0.5, colors='black',)
ax.vlines(x=-2, ymin=0, ymax=no_height_sim, lw=0.5, colors='black')
ax.vlines(x=-1, ymin=0, ymax=no_height_sim, lw=0.5, colors='black')
else:
N, bins, patches = ax.hist(df.loc[:, 'rt_resp'], bins=bins,
density=True, color='magenta', alpha=0.5)
for bin_size, bin, patch in zip(N, bins, patches):
if bin < 0:
plt.setp(patch, 'facecolor', 'cyan')
ax.hist(df_sim.loc[:, 'rt_resp'], bins=bins, density=True,
histtype='step', color='k', alpha=1, label=None)
ax.set_title('P(bias)={}'.format(round(df.loc[:, 'response'].mean(), 3),))
ax.set_xlabel('RT (s)')
ax.set_ylabel('Trials (prob. dens.)')
plt_nr += 1
# condition response plots:
ax = fig.add_subplot(nr_subjects,4,plt_nr)
df.loc[:,'rt_bin'] = pd.qcut(df['rt'], quantiles, labels=False)
d = df.groupby(['rt_bin']).mean().reset_index()
ax.errorbar(d.loc[:, "rt"], d.loc[:, "response"], fmt='-o', color='orange', markersize=10)
if df_sim is not None:
df_sim.loc[:,'rt_bin'] = pd.qcut(df_sim['rt'], quantiles, labels=False)
d = df_sim.groupby(['rt_bin']).mean().reset_index()
ax.errorbar(d.loc[:, "rt"], d.loc[:, "response"], fmt='x', color='k', markersize=6)
if xlim:
ax.set_xlim(xlim)
ax.set_ylim(0,1.25)
ax.set_title('Conditional response')
ax.set_xlabel('RT (quantiles)')
ax.set_ylabel('P(bias)')
plt_nr += 1
sns.despine(offset=3, trim=True)
plt.tight_layout()
return fig
Let’s simulate our own data, so we know what the fitting procedure should converge on:
# settings
go_nogo = True # should we put all RTs for one choice alternative to NaN (go-no data)?
n_subjects = 4
trials_per_level = 10000
# parameters:
params0 = {'cond':0, 'v':0.5, 'a':2.0, 't':0.3, 'z':0.5, 'dc':-0.2, 'sz':0, 'st':0, 'sv':0}
params1 = {'cond':1, 'v':0.5, 'a':2.0, 't':0.3, 'z':0.5, 'dc':0.2, 'sz':0, 'st':0, 'sv':0}
# simulate:
dfs = []
for i in range(n_subjects):
df0 = simulate_data(z=params0['z'], a=params0['a'], v=params0['v'], dc=params0['dc'],
t=params0['t'], sv=params0['sv'], st=params0['st'], sz=params0['sz'],
condition=params0['cond'], nr_trials1=trials_per_level, nr_trials2=trials_per_level)
df1 = simulate_data(z=params1['z'], a=params1['a'], v=params1['v'], dc=params1['dc'],
t=params1['t'], sv=params1['sv'], st=params1['st'], sz=params1['sz'],
condition=params1['cond'], nr_trials1=trials_per_level, nr_trials2=trials_per_level)
df = pd.concat((df0, df1))
df['subj_idx'] = i
dfs.append(df)
# combine in one dataframe:
df_emp = pd.concat(dfs)
if go_nogo:
df_emp.loc[df_emp["response"]==0, 'rt'] = np.NaN
df_emp
| rt | response | subj_idx | condition | correct | stimulus | |
|---|---|---|---|---|---|---|
| 0 | 0.880 | 1 | 0 | 0 | 1 | 1 |
| 1 | NaN | 0 | 0 | 0 | 0 | 1 |
| 2 | 1.499 | 1 | 0 | 0 | 1 | 1 |
| 3 | NaN | 0 | 0 | 0 | 0 | 1 |
| 4 | NaN | 0 | 0 | 0 | 0 | 1 |
| ... | ... | ... | ... | ... | ... | ... |
| 9995 | 0.829 | 1 | 3 | 1 | 0 | 0 |
| 9996 | 0.938 | 1 | 3 | 1 | 0 | 0 |
| 9997 | 1.674 | 1 | 3 | 1 | 0 | 0 |
| 9998 | NaN | 0 | 3 | 1 | 1 | 0 |
| 9999 | NaN | 0 | 3 | 1 | 1 | 0 |
160000 rows × 6 columns
Fit using the g-quare method.
# fit chi-square:
quantiles = [.1, .3, .5, .7, .9]
params_fitted = pd.concat(Parallel(n_jobs=n_subjects)(delayed(fit_subject)(data[1], quantiles)
for data in df_emp.groupby('subj_idx')))
params_fitted.drop(['bic', 'likelihood', 'penalty', 'z_trans(0)', 'z_trans(1)'], axis=1, inplace=True)
print(params_fitted)
a(0) a(1) v(0) v(1) t(0) t(1) z(0) 0 1.957684 1.997512 0.506885 0.503210 0.291217 0.304618 0.476925
1 1.992596 2.009814 0.495720 0.514267 0.286401 0.271875 0.481426
2 1.971131 1.996191 0.522817 0.495448 0.270259 0.283106 0.451006
3 1.999619 1.972885 0.501150 0.493823 0.309258 0.298655 0.512594
z(1) dc(0) dc(1)
0 0.503880 -0.157141 0.197714
1 0.470882 -0.158436 0.259854
2 0.478944 -0.096833 0.231788
3 0.489152 -0.223152 0.236227
# simulate data based on fitted params:
dfs = []
for i in range(n_subjects):
df0 = simulate_data(a=params_fitted.loc[i,'a(0)'], v=params_fitted.loc[i,'v(0)'],
t=params_fitted.loc[i,'t(0)'], z=params_fitted.loc[i,'z(0)'],
dc=params_fitted.loc[i,'dc(0)'], condition=0, nr_trials1=trials_per_level,
nr_trials2=trials_per_level)
df1 = simulate_data(a=params_fitted.loc[i,'a(1)'], v=params_fitted.loc[i,'v(1)'],
t=params_fitted.loc[i,'t(1)'], z=params_fitted.loc[i,'z(1)'],
dc=params_fitted.loc[i,'dc(1)'], condition=1, nr_trials1=trials_per_level,
nr_trials2=trials_per_level)
df = pd.concat((df0, df1))
df['subj_idx'] = i
dfs.append(df)
df_sim = pd.concat(dfs)
if go_nogo:
df_sim.loc[df_sim["response"]==0, 'rt'] = np.NaN
df_sim
| rt | response | subj_idx | condition | correct | stimulus | |
|---|---|---|---|---|---|---|
| 0 | 1.668217 | 1 | 0 | 0 | 1 | 1 |
| 1 | 1.870217 | 1 | 0 | 0 | 1 | 1 |
| 2 | 3.025217 | 1 | 0 | 0 | 1 | 1 |
| 3 | 0.914217 | 1 | 0 | 0 | 1 | 1 |
| 4 | 1.033217 | 1 | 0 | 0 | 1 | 1 |
| ... | ... | ... | ... | ... | ... | ... |
| 9995 | NaN | 0 | 3 | 1 | 1 | 0 |
| 9996 | NaN | 0 | 3 | 1 | 1 | 0 |
| 9997 | NaN | 0 | 3 | 1 | 1 | 0 |
| 9998 | 0.676655 | 1 | 3 | 1 | 0 | 0 |
| 9999 | 0.853655 | 1 | 3 | 1 | 0 | 0 |
160000 rows × 6 columns
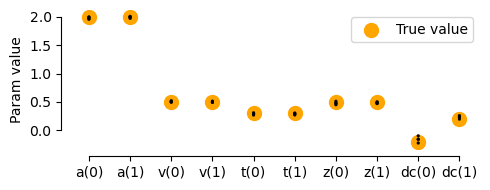
# Plot true vs recovered parameters:
x = np.arange(5) * 2
y0 = np.array([params0['a'], params0['v'], params0['t'], params0['z'], params0['dc']])
y1 = np.array([params1['a'], params1['v'], params1['t'], params1['z'], params1['dc']])
fig = plt.figure(figsize=(5,2))
ax = fig.add_subplot(111)
ax.scatter(x, y0, marker="o", s=100, color='orange', label='True value')
ax.scatter(x+1, y1, marker="o", s=100, color='orange',)
sns.stripplot(data=params_fitted, jitter=False, size=2, edgecolor='black', linewidth=0.25, alpha=1, palette=['black', 'black'], ax=ax)
plt.ylabel('Param value')
plt.legend()
sns.despine(offset=5, trim=True,)
plt.tight_layout()
plt.show()
# Plot data with model fit on top:
for c in np.unique(df_emp['condition']):
print('CONDITION {}'.format(c))
summary_plot(df_group=df_emp.loc[(df_emp['condition']==c),:],
df_sim_group=df_sim.loc[(df_emp['condition']==c),:])
plt.show()
CONDITION 0
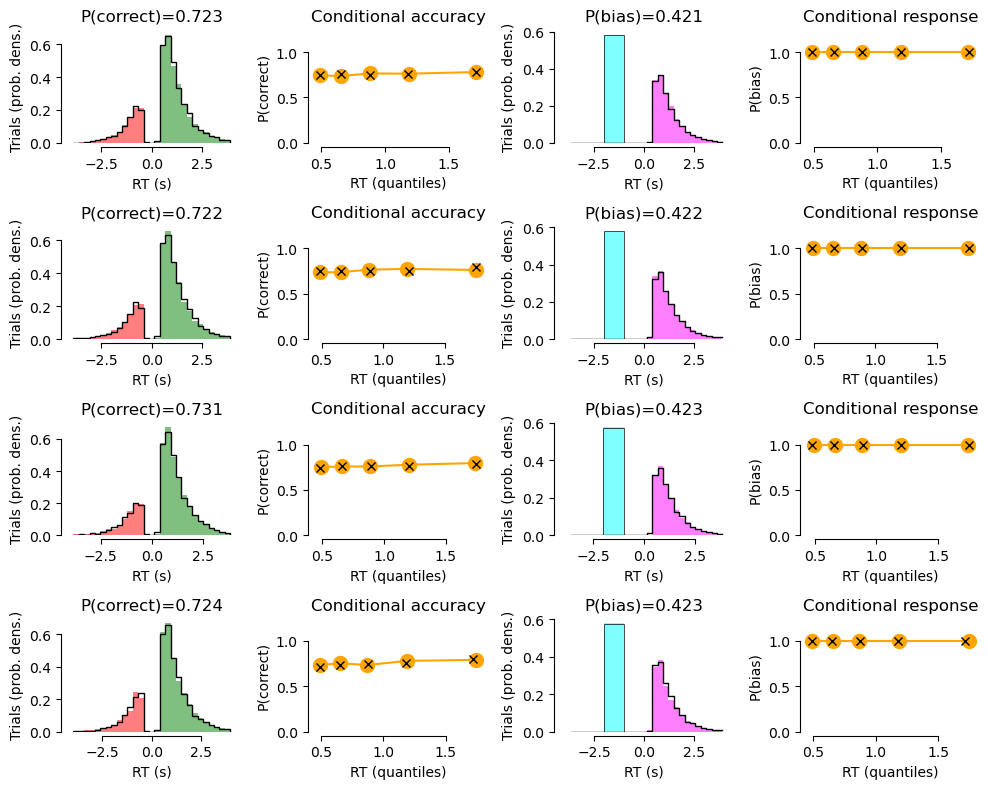
CONDITION 1