Tutorial on the “Model Plot”
The hddm.plotting._plot_func_model model, which we can supply to
hddm.plotting.plot_from_data or hddm.plotting.plot_posteriors
functions has become quite versatile (complex).
This tutorial is meant to illustrate some of it’s capabilities. We can generate didactically useful plots of a variety of generative models. The LAN Tutorial also shows some of the usecases.
Install (colab)
# package to help train networks
# !pip install git+https://github.com/AlexanderFengler/LANfactory
# package containing simulators for ssms
# !pip install git+https://github.com/AlexanderFengler/ssm_simulators
# packages related to hddm
# !pip install cython
# !pip install pymc==2.3.8
# !pip install git+https://github.com/hddm-devs/kabuki
# !pip install git+https://github.com/hddm-devs/hddm
Import modules
# MODULE IMPORTS ----
# warning settings
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
# Data management
import pandas as pd
import numpy as np
import pickle
# Plotting
import matplotlib.pyplot as plt
import matplotlib
import seaborn as sns
# Stats functionality
from statsmodels.distributions.empirical_distribution import ECDF
# HDDM
import hddm
from hddm.simulators.hddm_dataset_generators import simulator_h_c
Note a slight difference in how we treat generative models. Thanks to the LAN-extension, we have many generative models available to us, which we can broadbly classify as:
Legacy models, now titled:
ddm_hddm_baseandfull_ddm_hddm_base. Thefull_ddm_hddm_basemodel is used when any of thesv,storszparameters are positive.2-choice models enabled by the LAN-extension: amongst others the
levy,weibullandanglemodels.n-choice models enables by the LAN-extension: amongst others the
race_3andrace_4models.
The model plots work best with the 2-choice models enabled by the
LAN-extension. A n-choice version for the model plots exists (see
the _plot_func_model_n function) but is less well tested. Model
plots also work for the hddm_base models, however the positioning
of the reaction time and choice histograms is a little more tricky,
and will sometimes lead to unsatisfactory results.
Example 1: Simple Data from ddm_hddm_base model
from hddm.simulators.hddm_dataset_generators import simulator_h_c
model = 'ddm_hddm_base'
n_samples = 1000
data, full_parameter_dict = simulator_h_c(
n_subjects=1,
n_trials_per_subject=n_samples,
model=model,
p_outlier=0.00,
conditions=None,
depends_on=None,
regression_models=None,
regression_covariates=None,
group_only_regressors=False,
group_only=None,
fixed_at_default=None,
)
data
| rt | response | subj_idx | v | a | z | t | |
|---|---|---|---|---|---|---|---|
| 0 | 3.279359 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 1 | 2.632366 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 2 | 3.041361 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 3 | 2.648366 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 4 | 2.437368 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 995 | 2.478368 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 996 | 2.711365 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 997 | 3.343362 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 998 | 2.670366 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
| 999 | 2.893363 | 1.0 | 0 | 1.241863 | 2.258145 | 0.605863 | 2.262367 |
1000 rows × 7 columns
hddm.plotting.plot_from_data(df = data,
generative_model = model,
save = False,
make_transparent = False,
path = 'tmp_figures',
value_range = np.arange(-.1, 5, 0.1),
plot_func = hddm.plotting._plot_func_model,
keep_frame = True,
**{'hist_bottom': 0.})
plt.show()
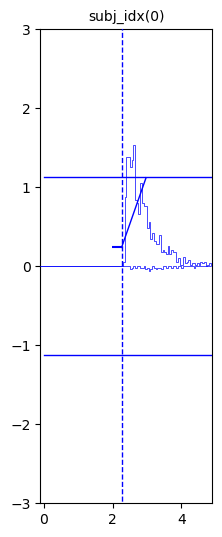
Example 2: Simple Data from a model enabled by LAN-extension
from hddm.simulators.hddm_dataset_generators import simulator_h_c
model = 'angle'
n_samples = 1000
data, full_parameter_dict = simulator_h_c(
n_subjects=1,
n_trials_per_subject=n_samples,
model=model,
p_outlier=0.00,
conditions=None,
depends_on=None,
regression_models=None,
regression_covariates=None,
group_only_regressors=False,
group_only=None,
fixed_at_default=None,
)
data
| rt | response | subj_idx | v | a | z | t | theta | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1.117624 | 1.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 1 | 2.047633 | 0.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 2 | 1.491619 | 1.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 3 | 1.583618 | 1.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 4 | 1.662617 | 1.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 995 | 1.283622 | 0.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 996 | 2.259643 | 1.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 997 | 1.866625 | 0.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 998 | 1.129624 | 1.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
| 999 | 2.129637 | 1.0 | 0 | 0.832015 | 1.953756 | 0.426032 | 0.685626 | 0.745569 |
1000 rows × 8 columns
hddm.plotting.plot_from_data(df = data,
generative_model = model,
save = False,
make_transparent = False,
path = 'tmp_figures',
value_range = np.arange(-.1, 5, 0.1),
plot_func = hddm.plotting._plot_func_model,
keep_frame = False,
**{'hist_bottom': 0})
plt.show()
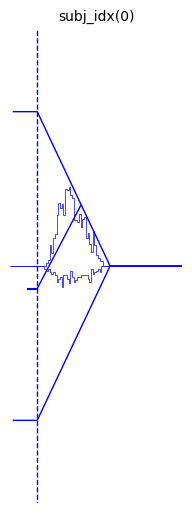
We can move around the histograms with the hist_bottom, argument
(kwarg).
hddm.plotting.plot_from_data(df = data,
generative_model = model,
save = False,
make_transparent = False,
path = 'tmp_figures',
value_range = np.arange(-.1, 5, 0.1),
plot_func = hddm.plotting._plot_func_model,
keep_frame = False,
**{'hist_bottom': data.a.values[0],
'ylim': 3})
plt.show()
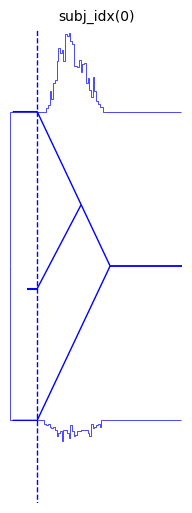
We can look at a few other arguments to illustrate the range of styling options.
hddm.plotting.plot_from_data(df = data,
generative_model = model,
save = False,
make_transparent = False,
path = 'tmp_figures',
value_range = np.arange(-.1, 7, 0.1),
plot_func = hddm.plotting._plot_func_model,
keep_frame = False,
figsize = (14, 6),
**{"hist_bottom": data.a.values[0],
"ylim": 3,
"add_trajectories": True,
"n_trajectories": 10,
"markersize_trajectory_rt_choice": 100,
"markertype_trajectory_rt_choice": "*",
"color_trajectories": {-1.:"red", 1.:"blue"},
"markercolor_trajectory_rt_choice": {-1.:"red",1.:'blue'},
"add_data_model_markersize_starting_point": 40,
"add_data_model_markertype_starting_point": '>',
"add_data_model_markershift_starting_point": -0.1,
"linewidth_histogram": 2,
"linewidth_model": 2,
"bin_size": 0.1,
"data_color": "black"})
plt.show()
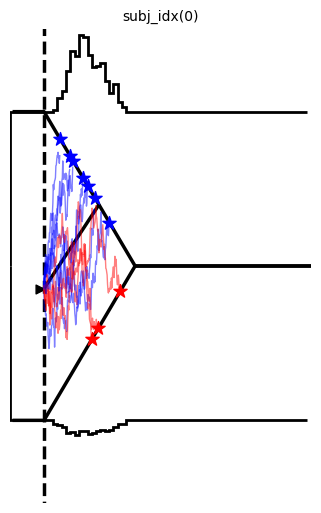
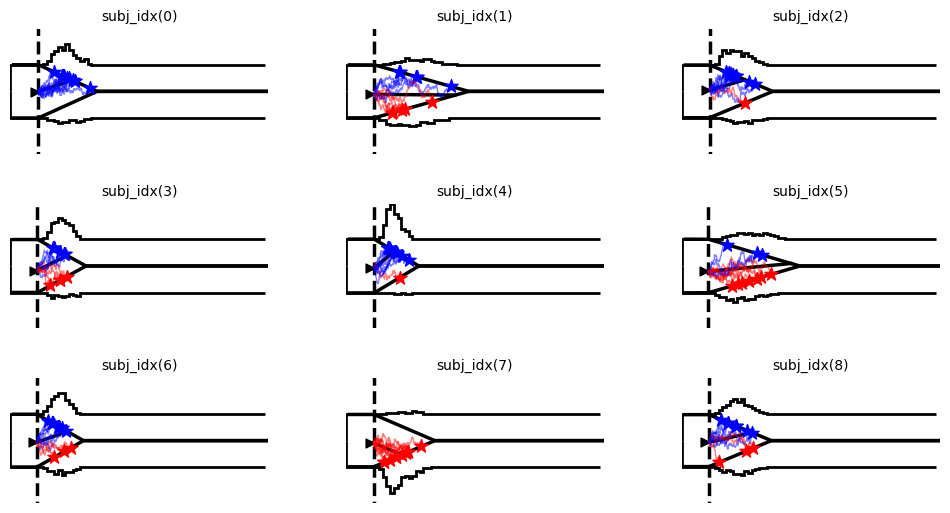
Example 3: More complex data
model = 'angle'
n_samples = 500
data, full_parameter_dict = simulator_h_c(
n_subjects=9,
n_trials_per_subject=n_samples,
model=model,
p_outlier=0.00,
conditions=None,
depends_on=None,
regression_models=None,
regression_covariates=None,
group_only_regressors=False,
group_only=None,
fixed_at_default=None,
)
hddm.plotting.plot_from_data(df = data,
generative_model = model,
save = False,
columns = 3,
make_transparent = False,
path = 'tmp_figures',
value_range = np.arange(-.1, 7, 0.1),
plot_func = hddm.plotting._plot_func_model,
keep_frame = False,
figsize = (12, 6),
**{"hist_bottom": data.a.values[0],
"ylim": 4,
"add_trajectories": True,
"n_trajectories": 10,
"markersize_trajectory_rt_choice": 100,
"markertype_trajectory_rt_choice": "*",
"color_trajectories": {-1.:"red", 1.:"blue"},
"markercolor_trajectory_rt_choice": {-1.:"red",1.:'blue'},
"add_data_model_markersize_starting_point": 40,
"add_data_model_markertype_starting_point": '>',
"add_data_model_markershift_starting_point": -0.1,
"linewidth_histogram": 2,
"linewidth_model": 2,
"bin_size": 0.1,
"data_color": "black"})
plt.show()
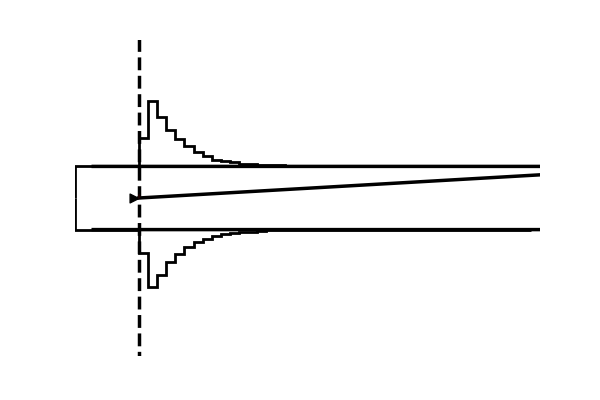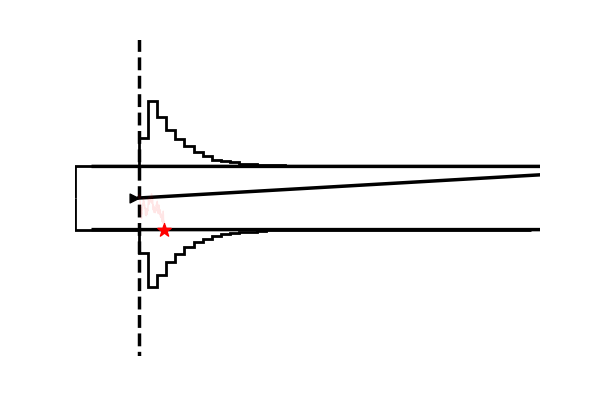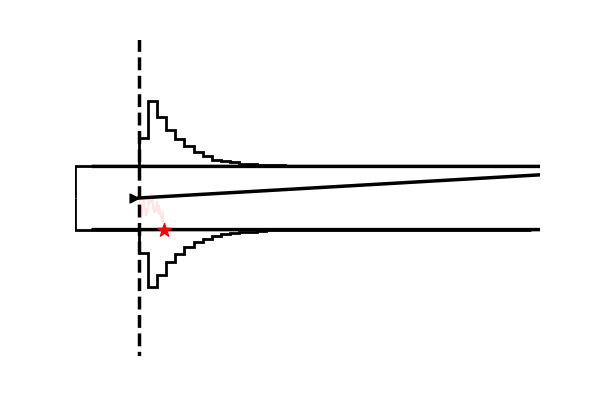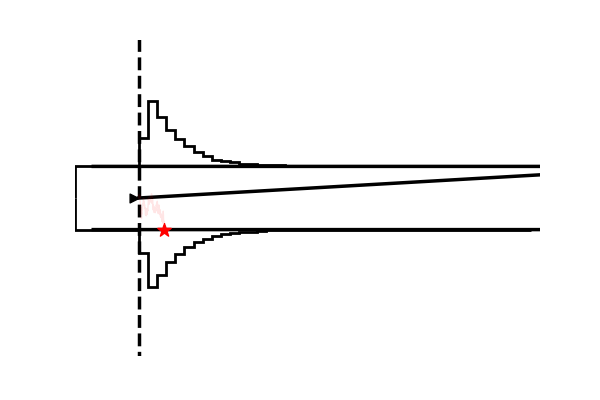
Example 4: Making gifs
Using the model plot, we can create fairly exciting gifs to illustrate the workings of various Sequential Sampling Models.
import os
import imageio # package for gifs
from hddm.simulators.basic_simulator import simulator
from hddm.simulators import hddm_preprocess
#
theta = np.array([[.1, .6, 0.5, .5]])
theta_secondary = np.array([-.3, 1., 0.5, 1.0])
model = 'ddm'
model_secondary = 'ddm'
n_samples=20000
n_samples_secondary=500 # If we want to have another reference dataset (can be useful for illustrating e.g. likelihood)
create_frames=True
make_gif=True
add_secondary_data=False
# Create trajectories (have to presimulate, then use for frame generation)
n_trajectories = 1
trajectories = []
trajectory_choices = []
for i in range(n_trajectories):
out_tmp = hddm.simulators.basic_simulator.simulator(theta = theta,
model = model,
n_samples = 1,
delta_t = 0.001)
trajectories.append(out_tmp[2]['trajectory'].T)
trajectory_choices.append(out_tmp[1].flatten())
trajectories_to_supply = np.concatenate(trajectories)
trajectory_choices_to_supply = np.concatenate(trajectory_choices)
trajectory_supply_dict = {'trajectories': trajectories_to_supply,
'trajectory_choices':trajectory_choices_to_supply}
# Create data
out = simulator(theta = theta,
model = model,
n_samples = n_samples,
delta_t = 0.001)
data = hddm_preprocess(out,
subj_id = '0',
add_model_parameters = True)
out_secondary = simulator(theta = theta_secondary,
model = model_secondary,
n_samples = n_samples_secondary,
delta_t = 0.001)
data_secondary = hddm_preprocess(out_secondary,
subj_id = '0',
add_model_parameters = True)
if create_frames:
frames = []
cnt = 0
for i in range(n_trajectories):
# Subset precomputed trajectories for current plot
tmp_trajectories = {}
tmp_trajectories['trajectories'] = trajectory_supply_dict['trajectories'][:(i + 1), :]
tmp_trajectories['trajectory_choices'] = trajectory_supply_dict['trajectory_choices'][:(i + 1)]
tmp_maxid = np.argmax(np.where(tmp_trajectories['trajectories'][i, :] > -999))
for j in range(10, tmp_maxid + 110, 10):
# Define all the plot options via a kwarg dict
plot_options_dict = {"alpha": 1.0, "ylim": 3.0,
"hist_bottom": data.a.values[0],
"add_data_rts": True, "add_data_model": True,
'add_trajectories': True,
"alpha_trajectories": 0.1,
'n_trajectories': None,
"supplied_trajectory": tmp_trajectories, #out[2]['trajectory'][:, 0][np.arange(0, out[2]['trajectory'][:, 0].shape[0], 10)],
"maxid_supplied_trajectory": j,
"color_trajectories": {-1.:"red", 1.:"blue"},
"data_color": "black",
"bin_size": 0.1,
"linewidth_histogram": 2,
"linewidth_model": 2,
"markersize_trajectory_rt_choice": 100,
"markertype_trajectory_rt_choice": "*",
"markercolor_trajectory_rt_choice": {-1.:"red",1.:'blue'},
"highlight_trajectory_rt_choice": True,
"add_data_model_keep_boundary": True,
"add_data_model_keep_slope": True,
"add_data_model_keep_ndt": True,
"add_data_model_keep_starting_point": True,
"add_data_model_markersize_starting_point": 40,
"add_data_model_markertype_starting_point": '>',
"add_data_model_markershift_starting_point": -0.05,
"secondary_data": None, #data_secondary,
"secondary_data_color": 'blue',
"secondary_data_label": None}
# Create plot
hddm.plotting.plot_from_data(
df=data,
generative_model=model,
save=True,
make_transparent = False,
path='tmp_figures',
save_name='myfig_' + str(cnt),
columns=1,
groupby=["subj_idx"],
figsize=(6, 4),
value_range=np.arange(-.2, 5, 0.1),
delta_t_model = 0.001,
keep_frame=False,
keep_title=False,
plot_func=hddm.plotting._plot_func_model,
**plot_options_dict)
plt.close('all')
# Read image, append to frames and delete it from disk
image = imageio.v2.imread('./tmp_figures/myfig_' + str(cnt) + '.png')
frames.append(image)
os.remove('./tmp_figures/myfig_' + str(cnt) + '.png')
cnt += 1
# Append a few frames with the last image to make the gif a bit better to view
for j in range(100):
frames.append(image)
imageio.mimsave('./tmp_gifs/example.gif',
frames,
fps = 50)
SegmentLocal